Jupyter notebooks are great for jobs like teaching, prototyping, and interactive documentation. In my former life as a postdoc I knew many skilled researchers who developed all their code in Jupyter notebooks, as it’s how they learned to code in the first place. Despite the user-friendly appearance, the raw files can be very complex. Let’s add a code cell to a notebook and see:
# Some data and a for loop
animals = ['lion','badger','dragon']
for animal in animals:
print(f"Oh no a {animal}!")Here is the cell in the raw .ipynb file:
{
"cell_type": "code",
"execution_count": null,
"id": "cdff1511-b4d0-4786-9451-9a8de61df394",
"metadata": {},
"outputs": [],
"source": [
"# Some data and a for loop\n",
"animals = ['lion','badger','dragon']\n",
"\n",
"for animal in animals:\n",
" print(f\"Oh no a {animal}!\")"
]
}Our short code cell is stored in a JSON format- easy to read if you're a machine, but not if you're a human. And this is just the code itself- if we actually run it and save the cell outputs (and all the metadata), a few code cells and a function definition could end up being hundreds of lines long. It gets worse- cell output images are stored as encoded strings. Here is a scatter plot with 4 data points:
{
"data": {
"image/png": "iVBORw0KGgoAAAANSUhEUgAAAiMAAAGdCAYAAADAAnMpAAAAOXRFWHRTb2Z0d2FyZQBNYX...",
"text/plain": [
"<Figure size 640x480 with 1 Axes>"
]
},
"metadata": {},
"output_type": "display_data"
}Note that this is the abridged version- the original string was over 11,000 characters long!
So what's the problem?
None of this is a problem if you are the only one who ever looks at your code, but this is becoming increasingly rare. Many journals now ask you to provide open-source code to reproduce your research, and some reviewers even run it to check it works! Git and Github are the industry standard method to provide open-source code, and are incredibly useful for jobs such as:
- Sharing code with others
- Tagging specific versions for papers or conference presentations
- Keeping track of changes (and tracking down the source of bugs!)
- Working collaboratively and getting code reviewed by a colleague
Some of these features are difficult or impossible if your code becomes a convoluted JSON filled with metadata. Imagine trying to review changes to the "image" above!
Furthermore, as your work becomes more complex you may wish to move things like function or class definitions into separate files. Generally these need to be standard .py or .jl files to use standard formatting and functionality. And if you want to speed up your calculations using advanced computing facilities like CSF or Bede, Jupyter is either difficult to work with or not supported at all.
Jupytext
Jupytext provides a solution, where standard Python files can be opened as an interactive notebook. Once installed, the interface is simple to access using Jupyterlab:
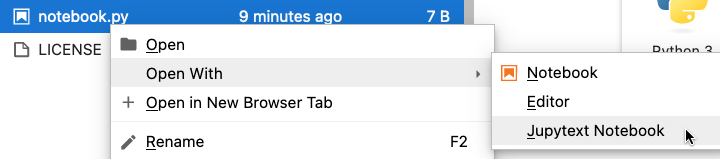
We can edit the notebook just like normal, but the code is stored in plain text, just like we see when writing it. This means Git will also see the changes in plain text. Standard Python/Julia files can also be formatted to display cells in your favourite IDE as well (mine is Spyder). The only real downside of this approach is that the cell outputs cannot be saved.
A second solution is to make paired files by running a short command from the terminal or command line. In Python:
jupytext --set-formats ipynb,py notebook.pyor in Julia:
jupytext --set-formats ipynb,jl notebook.ipynbThis gives us two linked files (one notebook, one standard Python/Julia file) with the same content. Changes in one file will be automatically mirrored by changes in the other. You can edit and save the .ipynb in the normal way (including the cell outputs), but only run version control on the .py/.jl file, which will automatically show your changes in plain text. So there you have it! Jupyter and Git, the best of friends.
Get in touch!
The Research Software Engineering (RSE) team are here to support researchers with their software engineering needs, including software development, coding training sessions, and data science/AI advice. If you would like an RSE to work with your research team and supply software development expertise, please get in touch by email or by attending one of our events, including our fortnightly informal drop-in sessions.

